About de.NBI ModSim
Resources, Services and Developments for Systems Biology Modeling.
de.NBI-ModSim is a partner project of de.NBI-SysBio node and has three major goals:
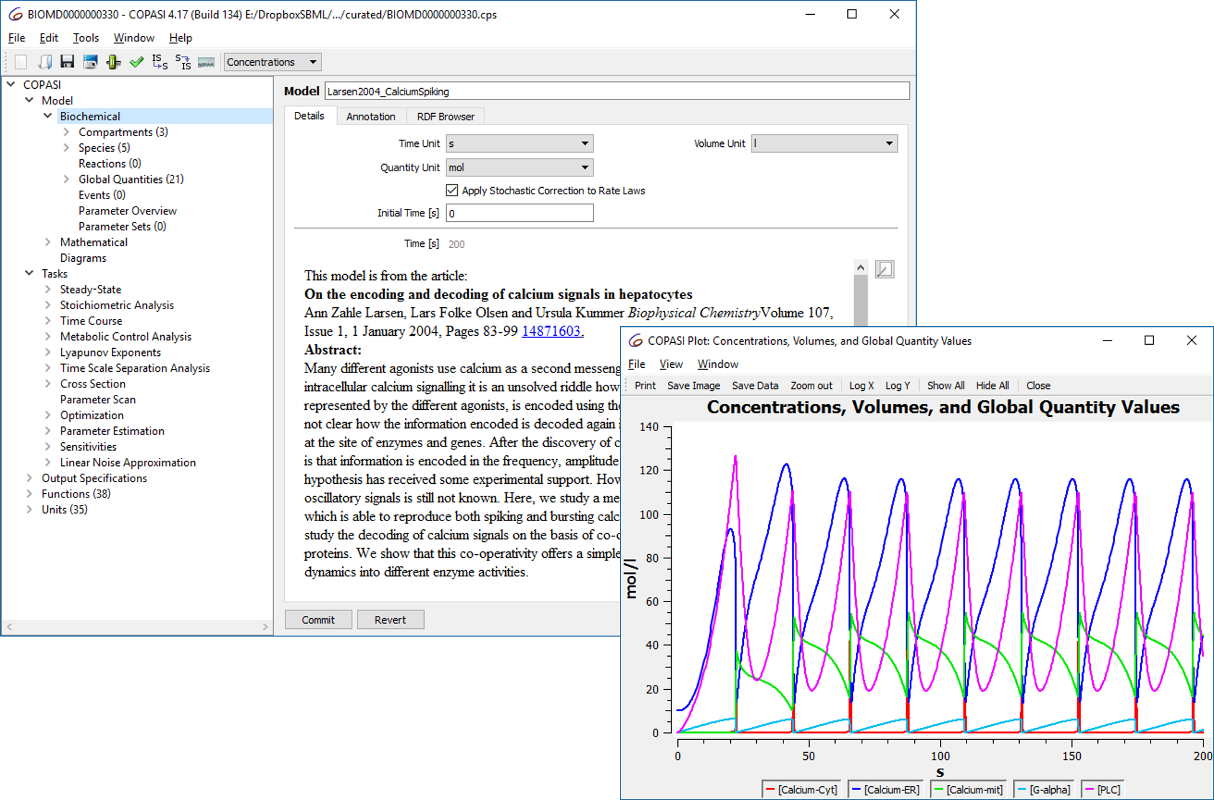
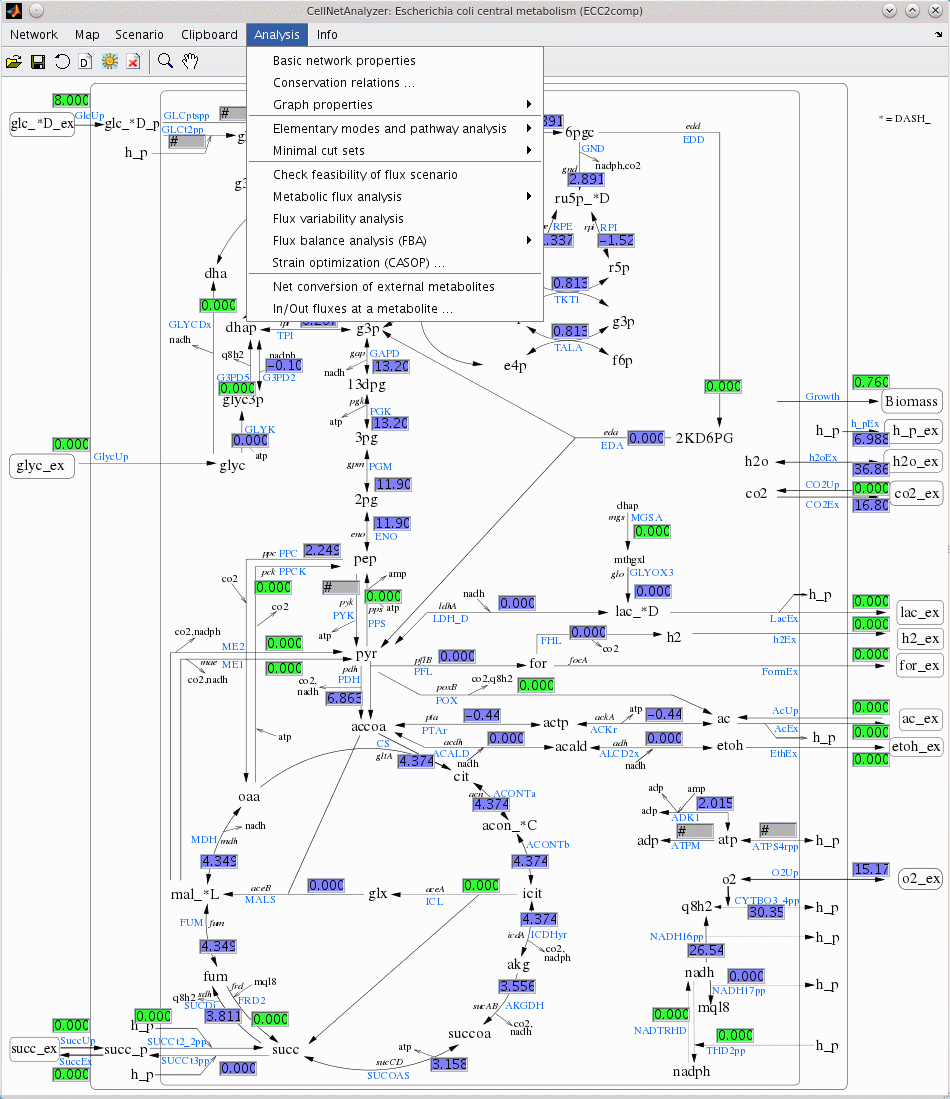
- Adding new resources to de.NBI: in particular, COPASI and CellNetAnalyzer, two complementary and frequently used software packages for modeling biological networks and systems will be promoted by de.NBI-ModSim.
- Providing services: extended user support and organization of workshops and training courses on mathematical modeling of biological systems including hands-on exercises with the software tools
- de.NBI-relevant developments: for example, integration of COPASI and CellNetAnalyzer with existing resources of de.NBI and make these tools compliant with model standardization and annotation schemes propagated by the de.NBI-SysBio node.
More about de.NBI